vec <- c(2.3,45.6,76,254.9)
vec[1] 2.3 45.6 76.0 254.9So, all the things that you manipulate in R, all the things that we encounter in R, are what might be called objects, objects can be all different kinds. They can contain all different kinds of data, but everything in R, is an object.
R has five basic atomic classes of objects, these are the very low level or basic classes of objects, and they are:
The most basic object is a vector:
In programming, a data structure is a format for organizing and storing data. Data structures are important to understand because you will work with them frequently when you use R for data analysis. The most common data structures in the R programming language include:
Think of a data structure like a house that contains your data.
There are two types of vectors: atomic vectors and lists. Coming up, you’ll learn about the basic properties of atomic vectors and lists, and how to use R code to create them.
First, we will go through the different types of atomic vectors. Then, you will learn how to use R code to create, identify, and name the vectors.
A vector is a group of data elements of the same type, stored in a sequence in R. You cannot have a vector that contains both logicals and numerics.
There are six primary types of atomic vectors: logical, integer, double, character (which contains strings), complex, and raw. The last two–complex and raw–aren’t as common in data analysis, so we will focus on the first four. Together, integer and double vectors are known as numeric vectors because they both contain numbers. This table summarizes the four primary types:
| Type | Description | Example |
|---|---|---|
| Logical | True/False | TRUE |
| Integer | Positive and negative whole values | 3 |
| Double | Decimal values | 101.175 |
| Character | String/character values | “Coding” |
This diagram illustrates the hierarchy of relationships among these four main types of vectors:
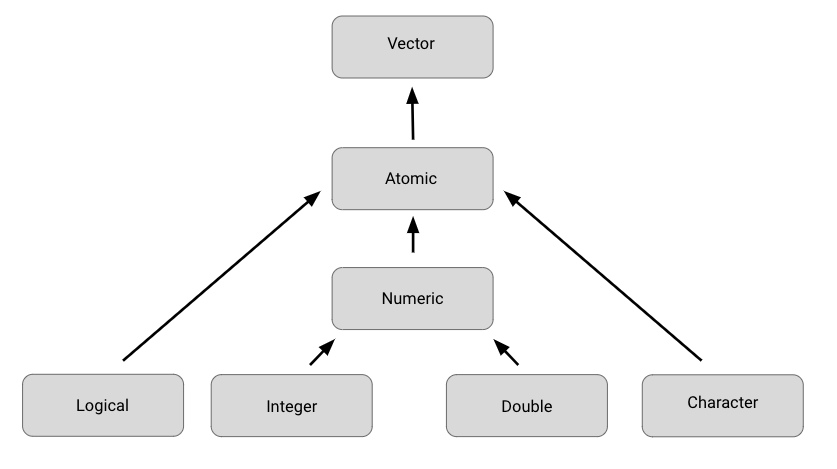
One way to create a vector is by using the c() function (called the “combine” function). The c() function in R combines multiple values into a vector. In R, this function is just the letter “c” followed by the values you want in your vector inside the parentheses, separated by a comma: c(x, y, z, …).
to combine multiple values together you use the c(x,y,z…)
For example, you can use the c() function to store numeric data in a vector.
c(2.5, 48.5, 101.5)to assign a vector to a variable we use
To create a vector of integers using the c() function, you must place the letter “L” directly after each number.
c(1L, 5L, 15L) #or x <- 1:45You can also create a vector containing characters or logicals.
c(“Sara” , “Lisa” , “Anna”) c(TRUE, FALSE, TRUE)Here is a use of as. as well as subset together. The second line starts with the inner subset(), so we are using r as data, and we are pulling the row where the column State to be equal to HI. That row is now a subset with however how many columns it has. From that subset we are pulling the value in column Hospital, then we are outputting the value as character.
as.character(subset(r, State == "HI")\$Hospital)As is used to specifically set the type of an element if you are unsure of it, or to change (coerce) it. Here we coerce all rows in column #11 to a numeric type regardless of their current type
outcome[ ,11] <- as.numeric(outcome[ ,11])What if we intentionally or by mistake mix classes when creating a vector? Well R will default to the least common denominator. Here are some examples:
You can intentionally/explicitly coerce a vector from one class to another using the as. function
So you see that x is an integer let’s coerce it into a numeric vector using as.
Now as you see it is a numeric, now let’s do coerce it as.logical
As you know 0=FALSE and( 1 and all >0 numbers = TRUE)
Every vector you create will have two key properties: type and length.You can determine what type of vector you are working with by using the typeof() function.
Place the code for the vector inside the parentheses of the function. When you run the function, R will tell you the type. For example:
typeof(c(“a” , “b”))
[1] "character"Notice that the output of the typeof function in this example is “character”. Similarly, if you use the typeof function on a vector with integer values, then the output will include “integer” instead:
typeof(c(1L , 3L))
[1] "integer"You can also check if a vector is a specific type by using an is function: is.logical(), is.double(), is.integer(), is.character(). In this example, R returns a value of TRUE because the vector contains integers.
x <- c(2L, 5L, 11L)
is.integer(x)
[1] TRUEIn this example, R returns a value of FALSE because the vector does not contain characters, rather it contains logicals.
y <- c(TRUE, TRUE, FALSE)
is.character(y)
[1] FALSEYou can determine the length of an existing vector–meaning the number of elements it contains–by using the length() function.
In this example, we use an assignment operator to assign the vector to the variable x. Then, we apply the length() function to the variable. When we run the function, R tells us the length is 3.
x <- c(33.5, 57.75, 120.05)
length(x)
[1] 3All types of vectors can be named. Names are useful for writing readable code and describing objects in R. You can name the elements of a vector with the names() function.
As an example, let’s assign the variable x to a new vector with three elements.
x <- c(1, 3, 5)You can use the names() function to assign a different name to each element of the vector.
names(x) <- c("a", "b", "c")Now, when you run the code, R shows that the first element of the vector is named a, the second b, and the third c with the corresponding values.
x
> a b c
> 1 3 5Remember that an atomic vector can only contain elements of the same type. If you want to store elements of different types in the same data structure, you can use a list. We’ll get to List later.
First, create a numeric vector num_vect that contains the values 0.5, 55, -10, and 6.
Now, create a variable called tf that gets the result of num_vect < 1, which is read as ‘num_vect is less than 1’, so we are basically asking if each element of the vector is < 1. I’m sure you can figure out what the answer should be
Try this without assignment
Both collapse and sep are arguments in the paste() function, we can use either, to set the separation value when we paste two objects together.
Create a character vector that contains the following words: “My”, “name”, “is”
my_char <- c("My","name","is")Paste all three together to join the elements with ONE space separation
paste(my_char, collapse = " ")
[1] "My name is"Now try
paste("Hello","world!",sep=" ")
[1] "Hello world!" Let’s add another element to the previous vector my_char by using the c() combine function again
my_name <- c(my_char, "santa")
paste(my_name, collapse = " ")
[1] "My name is santa"What do you think will happen if we paste a numeric with a character?
paste(c(1:3), c("X","Y","Z"), sep ="")
[1] "1X" "2Y" "3Z"Try paste(LETTERS, 1:4, sep = “-”), where LETTERS is a predefined variable in R containing a character vector of # all 26 letters in The English alphabet
paste(LETTERS, 1:4, sep = "-")
[1] "A-1" "B-2" "C-3" "D-4" "E-1" "F-2" "G-3" "H-4" "I-1" "J-2" "K-3" "L-4" "M-1" "N-2" "O-3" "P-4" "Q-1" "R-2" "S-3" "T-4" "U-1" "V-2" "W-3" "X-4" [25] "Y-1" "Z-2"What if we reverse the two elements?
paste( 1:4,LETTERS, sep = "-")
[1] "1-A" "2-B" "3-C" "4-D" "1-E" "2-F" "3-G" "4-H" "1-I" "2-J" "3-K" "4-L" "1-M" "2-N" "3-O" "4-P" "1-Q" "2-R" "3-S" "4-T" [21] "1-U" "2-V" "3-W" "4-X" "1-Y" "2-Z"tolower() changes all letters in column names to lower case for easier manipulation. Of course toupper() does the opposite
tolower(names(df))String splitter to separate characters we don’t want and then delete that column or rename it.
You can pull out a substring from a string. Let’s say I want to extract the first 6 characters of the string, I can use
substr("Jeffery is Santa's Helper", 1,6)
[1] "Jeffer"Lists are different from atomic vectors because their elements can be of any type—like dates, data frames, vectors, matrices, and more. Lists can even contain other lists.
You can create a list with the list() function. Similar to the c() function, the list() function is just list followed by the values you want in your list inside parentheses: list(x, y, z, …). In this example, we create a list that contains four different kinds of elements: character (“a”), integer (1L), double (1.5), and logical (TRUE).
list("a", 1L, 1.5, TRUE)Like we already mentioned, lists can contain other lists. If you want, you can even store a list inside a list inside a list—and so on.
list(list(list(1 , 3, 5)))structure(list): If you want to find out what types of elements a list contains, the structure of a list, you can use the str() function.
To do so, place the code for the list inside the parentheses of the function. When you run the function, R will display the data structure of the list by describing its elements and their types. Let’s apply the str() function to our first example of a list.
We run the function, then R tells us that the list contains four elements, and that the elements consist of four different types: character (chr), integer (int), number (num), and logical (logi).
Let’s use the str() function to discover the structure of our second example. First, let’s assign the list to the variable z to make it easier to input in the str() function.
List of 1
$ :List of 1
..$ :List of 3
.. ..$ : num 1
.. ..$ : num 3
.. ..$ : num 5The indentation of the $ symbols reflect the nested structure of this list. Here, there are three levels (so there is a list within a list within a list).
Lists, like vectors, can be named. You can name the elements of a list when you first create it with the list() function:
Tibbles are a little different from standard data frames. A data frame is a collection of columns, like a spreadsheet or a SQL table. Tibbles are like streamlined data frames that are automatically set to pull up only the first 10 rows of a dataset, and only as many columns as can fit on the screen.
Unlike data frames, tibbles never change the names of your variables, or the data types of your inputs. Overall, you can make more changes to data frames, but tibbles are easier to use. The tibble package is part of the core tidyverse. So, if you’ve already installed the tidyverse, you have what you need to start working with tibbles.
Now, let’s go through an example of how to create a tibble in R. You can use the pre-loaded diamonds dataset that’ preloaded into R. As a reminder, the diamonds dataset includes information about different diamond qualities, like carat, cut, color, clarity, and more. Tibbles can make printing easier. They also help you avoid overloading your console when working with large datasets. Tibbles are automatically set to only return the first ten rows of a dataset and as many columns as it can fit on the screen.
You can load the dataset with the data() function using the following code:
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsThen, let’s add the data frame to our data viewer in RStudio with the View() function.
View(diamonds)Now let’s create a tibble from the same dataset. You can create a tibble from existing data with the as_tibble() function. Indicate what data you’d like to use in the parentheses of the function. In this case, you will put the word “diamonds.”
# A tibble: 53,940 × 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
# ℹ 53,930 more rowsWhen you run the function, you get a tibble of the diamonds dataset.
Factor is a special type of vector, created to represent categorical data. Two types of factors exist:
One can think of a factor as an integer vector where each integer has a label. Think of an example as 1, 2, 3 where 1 is high, 2 is medium and 3 is low.
Factors are important because they are treated specially by modeling functions like lm() and glm() which are used for fitting linear models.
Factors with labels are more helpful, like Male/Female, Hot/Cold, because they are more easily understood than 1, 2 and 3 (in this case the user has to lookup the meaning of 1, 2,3 as opposed to named labels which are self-explanatory.
Think of this as storing data that have labels that are categorical but have no ordering. For example: Male/Female.
Represent things that are ranked. They have an order, but they are not numerical. For example, in a university you have professors, associate professors, assistant professors. Those are categorical but they are ordered.
I can count how many of each
If we want to strip out the class we can
Here we can set the levels manually
A matrix is a two-dimensional collection of data elements. This means it has both rows and columns. By contrast, a vector is a one-dimensional sequence of data elements. But like vectors, matrices can only contain a single data type. For example, you can’t have both logicals and numerics in a matrix.
We’ll cover matrices and data frames. Both represent ‘rectangular’ data types, meaning that they are used to store tabular data, with rows and columns. The main difference, as you’ll see, is that matrices can only contain a single class of data, while data frames can consist of many different classes of data.
To create a matrix in R, you can use the matrix() function. The matrix() function has two main arguments that you enter in the parentheses.
[,1] [,2] [,3]
[1,] NA NA NA
[2,] NA NA NA[1] 2 3$dim
[1] 2 3For example, imagine you want to create a 2x3 (two rows by three columns) matrix containing the values 3-8. - First, enter a vector containing that series of numbers: c(3:8). - Then, enter a comma. Finally, enter nrow = 2 to specify the number of rows.
The million dollar question you should be asking now is: How does R know it is supposed to be a 2x3 since all that was given was nrow=2?
Well if you count the objects in the vector (3:8) you come up with 6, and since we set nrows =2 then R automatically places half of the 6 objects in one row and the other half in the second row.
I’m sure you are asking yourself, what if we have 7 objects in the vector and we specify nrow=2 what does R do? Well, here is what it does: It not only tries to create a matrix for you by repeating 3 in the last empty cell, it also gives a warning.
Of course, if you are in a situation that you are too lazy to figure out if the vector you are trying to create a matrix from is divisible into the nrow value or not, you can always use this matrix() function and you’ll find out soon enough, or you can use another method we’ll get to.
Warning in matrix(c(3:9), nrow = 2): data length [7] is not a sub-multiple or
multiple of the number of rows [2] [,1] [,2] [,3] [,4]
[1,] 3 5 7 9
[2,] 4 6 8 3You can also choose to specify the number of columns (ncol = ) instead of the number of rows (nrow = ).
Here I am assigning a value to the dim attribute of m. I’m saying take this vector and transform it into a matrix
Let me break it down in pieces as to how we get a matrix out of the above:
Another way to create a matrix, and this is a common way, is by binding columns or binding rows.
So for example, suppose I have the two objects x, which is sequenced from 1 to 3 and y, which is a sequence from 10 to 12.
If I cbind those two objects, then I’ll get a matrix where the first column is 1 to 3, and the second column is 10 to 12.
So this is kind of what you might expect would happen.
If I rbind those two objects, then the first row will be 1 to 3, and the second row will be 10 through 12.
So cbind-ing and rbind–ing is another way to create a matrix.
What if we give the vector a dim attribute?
The dim() function allows you to get OR set the `dim` attribute for an R object. In this case, we assigned the value c(4, 5) to the `dim` attribute of my_vector like we did in the example above
Another way is to call the attributes function on my_vector and we get the (rows,columns)
Now that we have assigned a dimension to it with 4 rows and 5 columns we have a matrix not a vector any more, let’s see what we have now. As you see we have a matrix comprised of all the 20 numbers in 4 rows and 5 columns. Now let’s confirm we have a matrix with class()
What if we want to add a label for each row. In other word, let’s pretend each row represents the records of one patient, and so we want to have one column that contains the name of each patient for each row.
Let’s start by creating a character vector containing the name of our patients.
Now we’ll column bind the new names column to the existing matrix using cbind
patients
[1,] "Bill" "1" "5" "9" "13" "17"
[2,] "Gina" "2" "6" "10" "14" "18"
[3,] "Kelly" "3" "7" "11" "15" "19"
[4,] "Sean" "4" "8" "12" "16" "20"OH OH it appears that we ended up with a matrix of characters not numbers, that’s implicit coercion
So, we’re still left with the question of how to include the names of our patients in the table without destroying the integrity of our numeric data. Try the following:
Data frames are the most common way of storing and analyzing data in R, so it’s important to understand what they are and how to create them.
A data frame is a collection of columns–similar to a spreadsheet or SQL table. Each column has a name at the top that represents a variable, and includes one observation per row. Data frames help summarize data and organize it into a format that is easy to read and use. Matrices have to be of the same type but DF can have different types in each column.
For example, the data frame below shows the “diamonds” dataset, which is one of the preloaded datasets in R. Each column contains a single variable that is related to diamonds: carat, cut, color, clarity, depth, and so on. Each row represents a single observation.
There are a few key things to keep in mind when you are working with data frames:
If you need to manually create a data frame in R, you can use the data.frame() function.
The data.frame() function takes any number of arguments and returns a single object of class `data.frame` that is composed of the original objects.
It’s also possible to assign names to the individual rows and columns of a data frame, which presents another possible way of determining which row of values in our table belongs to each patient. But since we already solved that issue we can skip it. Instead of using cbind like we did just a few lines up, we just use data.frame() and we get:
Let’s say we now want to label or assign a name to each column in our df. First create a vector containing one element for each column and then use colnames() function to set the colnames for our df.
This is similar to setting the dim() earlier:
patient age weight bp rating test
1 Bill 1 5 9 13 17
2 Gina 2 6 10 14 18
3 Kelly 3 7 11 15 19
4 Sean 4 8 12 16 20data.frame - more
The data.frame() function takes vectors as input. In the parentheses, enter the name of the column, followed by an equals sign, and then the vector you want to input for that column. In this example, the x column is a vector with elements 1, 2, 3, and the y column is a vector with elements 1.5, 5.5, 7.5.
x y
1 1 1.5
2 2 5.5
3 3 7.5In most cases, you won’t need to manually create a data frame yourself, as you will typically import data from another source, such as a .csv file, a relational database, or a software program.
head() function returns the columns and the first several rows of data.
Another way to summarize the data frame
glimpse(all_trips19_20)
Rows: 119,390
Columns: 32
$ hotel <chr> "Resort Hotel", "Resort Hotel", "Resor…
$ is_canceled <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0,…
$ lead_time <dbl> 342, 737, 7, 13, 14, 14, 0, 9, 85, 75,…
$ arrival_date_year <dbl> 2015, 2015, 2015, 2015, 2015, 2015, 20…
$ arrival_date_month <chr> "July", "July", "July", "July", "July"… ...Lists all the column names
colnames(df)names <- c("Yasha", "Shaya", "Anjie", "Coyote")
age <- c(7,2 ,7 ,5 )
people <- data.frame(names, age) #call it people
head (people) #preview names age
1 Yasha 7
2 Shaya 2
3 Anjie 7
4 Coyote 5'data.frame': 4 obs. of 2 variables:
$ names: chr "Yasha" "Shaya" "Anjie" "Coyote"
$ age : num 7 2 7 5Rows: 4
Columns: 2
$ names <chr> "Yasha", "Shaya", "Anjie", "Coyote"
$ age <dbl> 7, 2, 7, 5[1] "names" "age" names age age_in_20
1 Yasha 7 27
2 Shaya 2 22
3 Anjie 7 27
4 Coyote 5 25